
————————————————————
Species distribution modeling,
calibration and evaluation,
ensemble modeling
————————————————————
Installation
-
Stable version
from cran :
install.packages("biomod2", dependencies = TRUE)-
Development version
from biomodhub :
library(devtools)
devtools::install_github("biomodhub/biomod2", dependencies = TRUE)
All changes between versions are detailed in News.
A new video tutorial is available on the Video page.
biomod 4.3-4-6 - Abundance modeling
Please feel free to indicate if you notice some strange new behaviors !
What is changed ?
- Nothing for presence/absence (or presence-only) modelling.
-
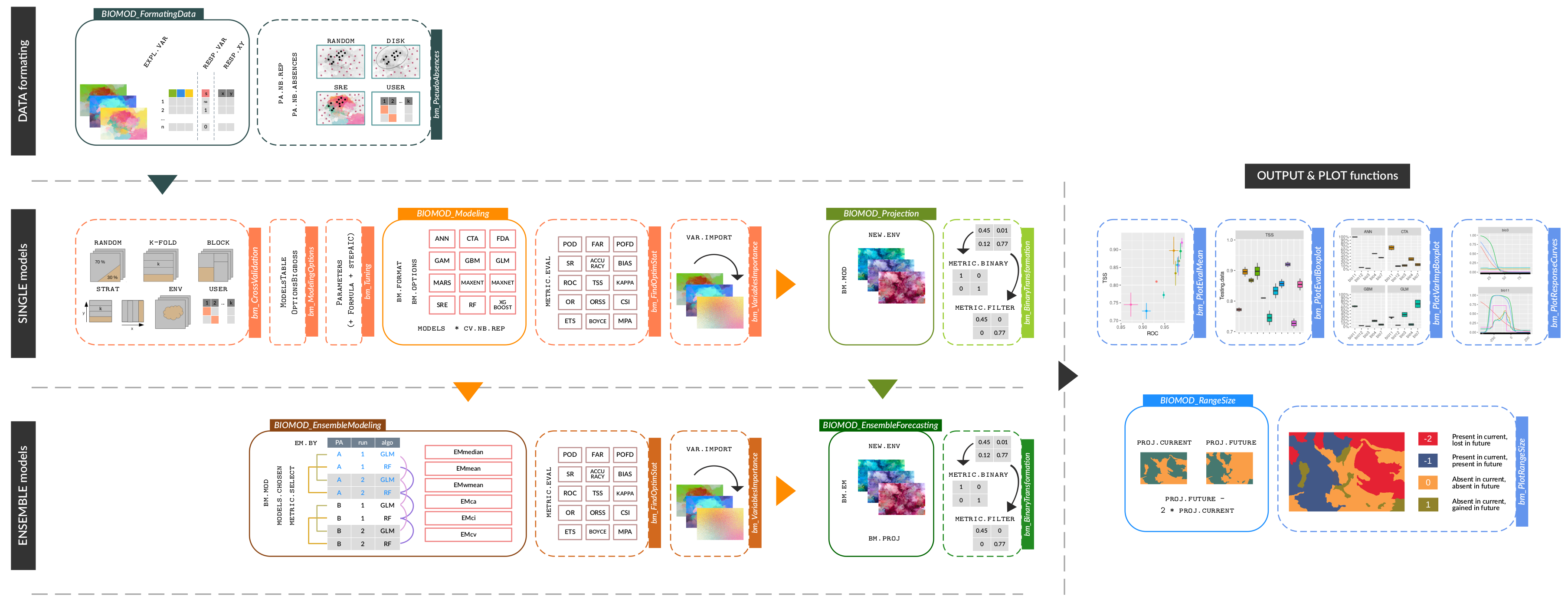
bm_RangeSizecomputes the difference between binary predictions,
and is called byBIOMOD_RangeSizewhich can take as inputBIOMOD.projection.outobjects. - Vignettes and example tutorials have been updated.
What is new ?
- Non-binary data ! Check the Main functions (abund) page for examples.
- 6 values can be given to
data.typenew parameter inBIOMOD_FormatingData:
binary, count, multiclass, ordinal, relative, abundance. - 2 new ensemble models have been added to deal with qualitative data type :
EMmodeandEMfreq.
-
DNN (Deep Neural Network) model has been added through
citopackage. It can be used with all data types.
Be sure to have a look at cito documentation before, especially the part about thetorchinstallation.
-
bm_ModelAnalysisrenders classical graphics to explore the quality of single models -
BIOMOD_Reportrenders.htmlreports to help summarize modeling steps
biomod 4.2-6 - Improved OptionsBigBoss and new model
What is changed ?
- To improve the models, we made some change in the options for
OptionsBigboss. (This only affects the ANN, CTA and RF models.) You can check all your options with theget_options()function. - To reduce the tuning calculation time, we update the tuning ranges for ANN, FDA and MARS models.
What is new ?
-
biomod2has a new model: RFd. It’s a Random Forest model with a down-sampling method. - You can now define seed.val for
bm_PseudoAbsences()andBIOMOD_FormatingData(). - New fact.aggr argument, for pseudo-absences selection with the random and disk methods, allows to reduce the resolution of the environment.
- Possibility to give the same options for all datasets with “for_all_datasets” in
bm_ModelingOptions().
biomod 4.2-5 - Modeling options & Tuning Update
What is changed ?
- modeling options are now automatically retrieved from single models functions, normally allowing the use of all arguments taken into account by these functions
- tuning has been cleaned up, but keep in mind that it is still a quite long running process
- in consequence,
BIOMOD_ModelingOptionsandBIOMOD_Tuningfunctions become secondary functions (bm_ModelingOptionsandbm_Tuning), and modeling options can be directly built throughBIOMOD_Modelingfunction
What is new ?
-
ModelsTableandOptionsBigbossdatasets (note that improvement of bigboss modeling options is planned in near future) - 3 new vignettes have been created :
- data preparation (questions you should ask yourself before modeling)
- cross-validation (to prepare your own calibration / validation datasets)
- modeling options (to help you navigate through the new way of parametrizing single models)
biomod 4.2 - Terra Update
What is changed ?
-
biomod2now relies on the newterrapackage that aims at replacingrasterandsp. -
biomod2is still compatible with old format such asRasterStackandSpatialPointsDataFrame. -
biomod2function will sometimes returnSpatRasterfrom packageterrathat you can always convert intoRasterStackusing functionstackinraster.
biomod 4.1 is now available
/!\ Package fresh start… meaning some changes in function names and parameters. We apologize for the trouble >{o.o}<
Sorry for the inconvenience, and please feel free to indicate if you notice some strange new behaviors !
What is changed ?
- all code functions have been cleaned, and old / unused functions have been removed
- function names have been standardized (
BIOMOD_for main functions,bm_for secondary functions) - parameter names have been standardized (same typo, same names for similar parameters across functions)
- all documentation and examples have been cleaned up
What is new ?
- plot functions have been re-written with
ggplot2 -
biomod2website has been created, with properroxygen2documentation and help vignettes
